Genome #4: The state of Covid-19 genomic surveillance in India - Part 3
The time to ramp up our genomic surveillance is now. Increased Private partnership might be a quick and stable fix.
This post is Part 3 of a three-part series. In Part 1 of this series, we looked at how India’s genomic surveillance has been abysmally suboptimal, both at the national and state level, with a huge state-to-state variation. Part 2 explored the variation in deposited samples across states. In this part, we look into the details of our sequencing potential and why we should not be shying away from involving the Private sector in enhancing our surveillance infrastructure.
The Omicron SARS-CoV-2 variant (B.1.1.529) was first reported by researchers in Bostwana and South Africa on 24th November 2021. On 26th November, World Health Organization (WHO) flagged it as a Variant of Concern which essentially means that it is associated with one or more of the following a) increased transmissibility b) increased severity or change in the clinical presentation of the disease c) decreased effectiveness of existing public health measures, diagnostics, vaccines, therapeutics.

Why is the Omicron concerning?
The Omicron variant has around 30+ mutations in its spike protein, some of which are concerning. Spike protein is a component on the surface of the virus that allows it to bind to human cells. The antibodies produced by the immune system to fight against the virus in our body post Covid-19 infection, chiefly target the spike protein . The presence of a large number of mutations in its spike protein gives the Omicron an advantage - it can evade the antibodies produced by previous infection, vaccination or even monoclonal antibodies. For example, a recent notification from Regeneron hints that its antibody cocktail might not be as effective against the Omicron variant:
To date, there have been no direct data testing the Omicron variant’s resistance to vaccineinduced and monoclonal antibody-conveyed immunity. Prior in vitro analyses and structural modeling regarding the individual mutations present in the Omicron variant indicate that there may be reduced neutralization activity of both vaccine-induced and monoclonal antibodyconveyed immunity, including the current REGEN-COV antibodies. Further analyses are ongoing to confirm and quantify this potential impact using the actual Omicron variant sequence.
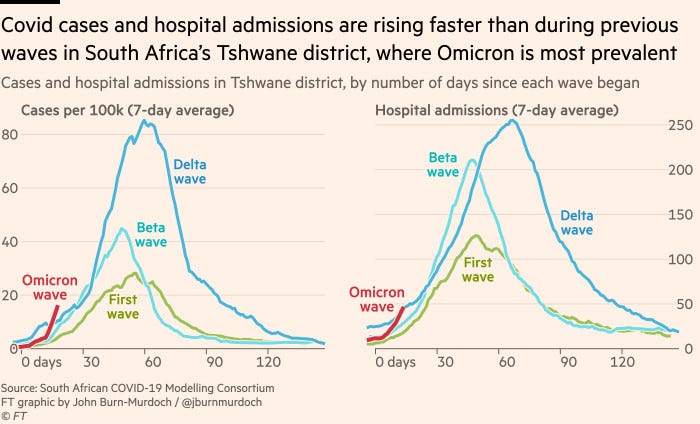
Twitter is full of threads on Micron and what it means for us and I will link a few at the end of this post. We do not have enough information to conclude anything at this point, but to summarize - we have good reasons to be concerned. The plot below, from a recent Financial Times article, shows that both cases and hospital admissions are steeply rising (too early to do any meaningful comparisons, but the rate is currently higher than delta):
What should be India’s strategy?
Omicron has already made its way into India, with the current count at 20. While our adult vaccination percentage lingers around half-century, we need to be monitoring the situation very carefully.
The first step of monitoring, besides continuing to vaccinate the eligible population, is to detect Omicron cases. While Indian labs are working on targeted RT-PCR tests for detecting Omicron variants, it might be currently possible to “guess” from a RT-PCR test (depending on the testing kit) if the sample is likely Omicron variant. This “guess” can then be confirmed via genomic sequencing. We look at how such a testing strategy would work.
How to guess an Omicron variant from an “eligible” RT-PCR test?
An RT-PCR test relies on a special set of sequences called the primer sequences, that can recognize some part of the known genes of the SARS-CoV-2 virus. A positive RT-PCR test indicates if starting from these primary sequences, it was able to find the presence of any of the SARS-CoV-2 genes that its primers are designed to detect.

Most RT-PCR tests are designed to detect one or multiple regions from one or more of the following genes: RNA-dependent RNA polymerase (RdRp), envelope (E) and nucleocapsid (N) or the Spike (S) gene. Since the RT-PCR kit has primers that were designed after studying the sequence of the previous mutants of these genes, a new variant with a large number of mutations in a gene the primers target might lead to a “target failure” of that gene.
An RT-PCR kit designed with primers against the E, N, and S genes will result in S-gene target failure (SGTF) for the Omicron variant and can be used as a first pass test before doing a genomic-sequencing based confirmatory test. However, a SGTF does not necessarily imply that the variant is Omicron, as any other set of mutations in the S gene (different from those in Omicron) can also cause the default primers in the kit to not work.
Can the RT-PCR kits in use in India detect S gene dropout?
Short answer, NO.
Most kits used in India appear to target the E, N and RdRp gene. Quoting an article from Business Standard:
"Most of the current ICMR approved RT-PCR kits being used in India target the E, Rd Rp and N genes. The mutations in the latest variant have occurred in the S gene. The common RT-PCR kits being used will be able to identify positive or negative, but will not be able to identify if the positive result is due to the mutation in the S gene," Arjun Dang, CEO, Dr Dangs Lab
I decided to confirm Dr. Dangs’ claim by doing a quick scan of the list of approved kits by ICMR and CDSCO. None of the lists however provide details on which genes are tested. Since ICMR actually tested these kits, it would have been useful to have this documented during the process of testing. Cataloging the gene details of each kit is a tedious process since it requires going through each kits’ manual and doing a quick scan of the list of genes its primers are designed to target. I started doing this for a few of the kits on this Google spreadsheet (Figure 5), hoping this would probably help someone decide to procure kits that have S-gene primers.
The fact that the majority of RT PCR kits that are currently in use in India cannot detect S genes implies that we cannot directly rely on our current testing procedure to inform us about Omicron prevalence. One obvious solution to this problem is to move to more robust RT-PCR kits that use a bigger pool of genes, such as TaqPath. Maharashtra and Karnataka have already started mass procuring the Taqpath kits.

A viable solution to properly detect Omicron prevalence in India is to ramp up genomic surveillance. The Government of India decided to set up the Indian SARS-CoV-2 Consortium on Genomics (INSACOG) on 25th December 2020 to increase the genomic surveillance for Covid-19 in the country. But how far have we come in one year?
How is India’s genomic surveillance doing?
In Part one and Part two of this series, we saw how India’s genomic surveillance was lagging behind its target of sequencing 5% of the samples testing positive. We also saw the states had huge disparities in the number of samples deposited to GISAID, a public repository where all the countries upload Covid-19 genomic sequence. This keeps the rest of the world informed about how different variants are evolving and can be the first step in making an informed policy decision.
India is still way below its 5% target
We are still not sequencing enough genomes (Figure 6). The definition of “enough” here is slightly arbitrary, but a study recommends sequencing at least 5% of the samples and INSACOG was established with the same aim:
It is proposed that 5% of the positive specimens (the representative number from each district/State to be decided by the CSU) detected daily will be referred to the designated RGSL for genome sequencing.
A recent report in TheWire found that the number of samples collected for sequencing dropped after June 2021 and halved through July and August 2021. We can look at the GISAID data and see that it is indeed the trend on a daily basis (Figure 5 and Figure 6). However, it is incorrect to focus on absolute numbers here, given the total cases had also started declining following June 2021.


The above two figures will probably mislead the reader to think that India’s sequencing efforts have taken a toll after April 2021. However, in these settings, the absolute numbers are largely not so useful. What we are interested in is how many of the samples testing positive were sequenced (and deposited to GISAID), i.e. the proportion of samples that are being sequenced. While that proportion has been abysmally low than the desired target of 5%, India’s efforts towards sequencing seem to be more or less unperturbed (Figure 7). We have been sequencing <1% of the samples throughout 2021 and this number has revoled around 0.5% between July - October 2021. Median delay before the samples are collected and before they are deposited to GISAID is around 70 days and as such the estimates in Figure 7 for the last 3 months are unreliable (which can partially explain the decline we observe during October 2021 - December 2021).

We still have a huge state-to-state disparity
We can look at how each state has been contributing towards the national sequencing efforts. Telangana ranks the highest with 2.36% median sequences shared with GISAID between March 2020 and November 2021, followed by Gujarat with 0.93% and Mizoram (0.89%).

Bihar, Uttar Pradesh, and Tamilnadu rank at the bottom of the list proportion of sequences shared over the course of the entire pandemic while the north-eastern states seem to have overall good surveillance, particularly after the second wave when these states particularly witnessed an uptick in cases. Kerala remains an exception which we discussed in Part one.

Only a few labs are doing the heavy lifting
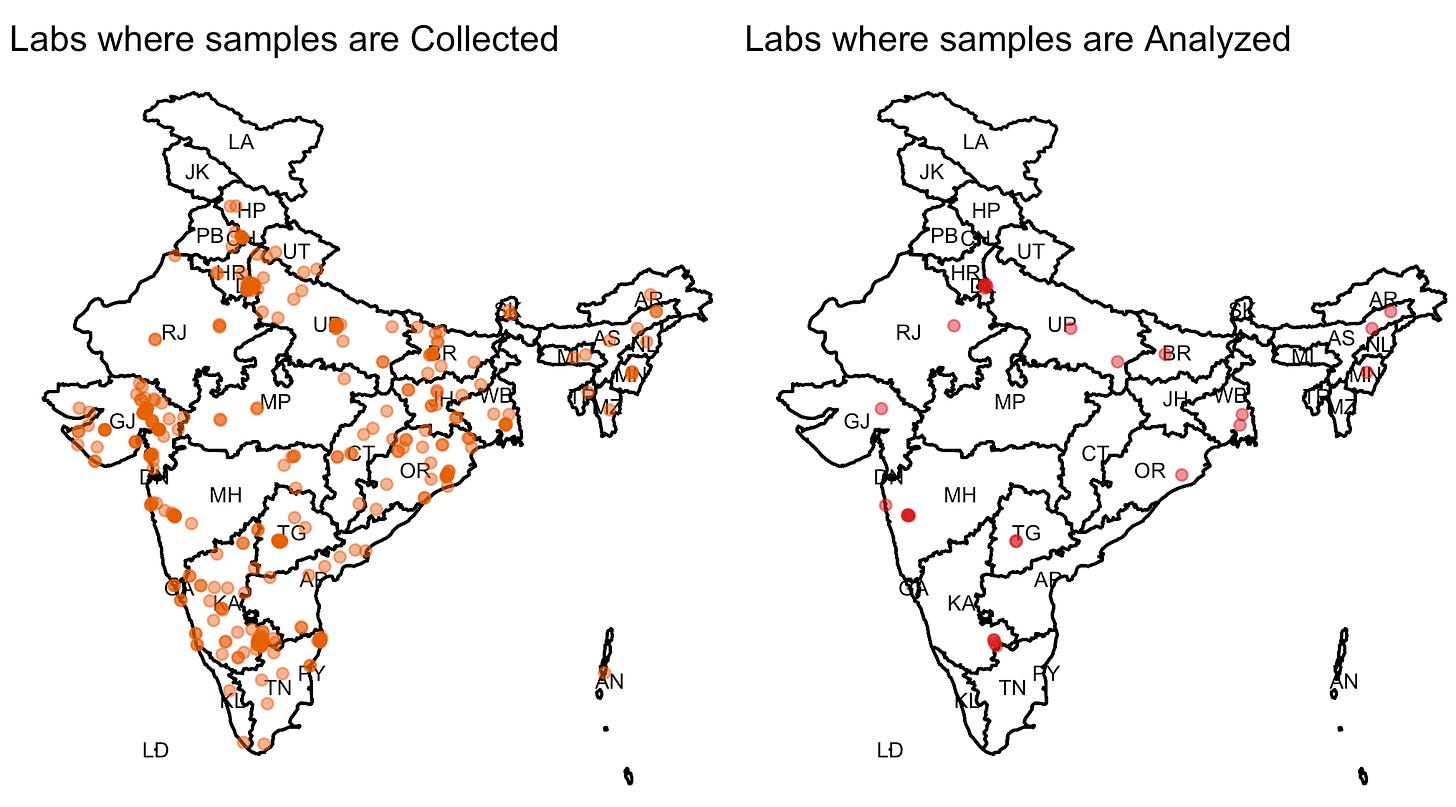
To start with, INSACOG was a consortium of 10 labs that was later expanded to incorporate 18 more labs. Samples are collected at “sentinel” sites which then get shipped to the responsible regional lab for sequencing. I downloaded the metadata from GISAID and mapped the sentinel sites after retrieving their latitude/longitude using Google maps API. For some reason J&K and Ladakh labs are missing because of the API’s failure to retrieve accurate coordinates for these. Karnataka has the highest sentinel sites (186) followed by Gujarat (88) and Delhi (43). Mizoram, Nagaland, and Tripura appear to have only one sentinel site. Maharashtra has the highest number of labs (6) that analyze the sequences with 5 of these in Pune itself. Delhi and Karnataka both have four labs each, while Bihar, Gujarat, Manipur, Odisha, and Rajasthan have one lab each.
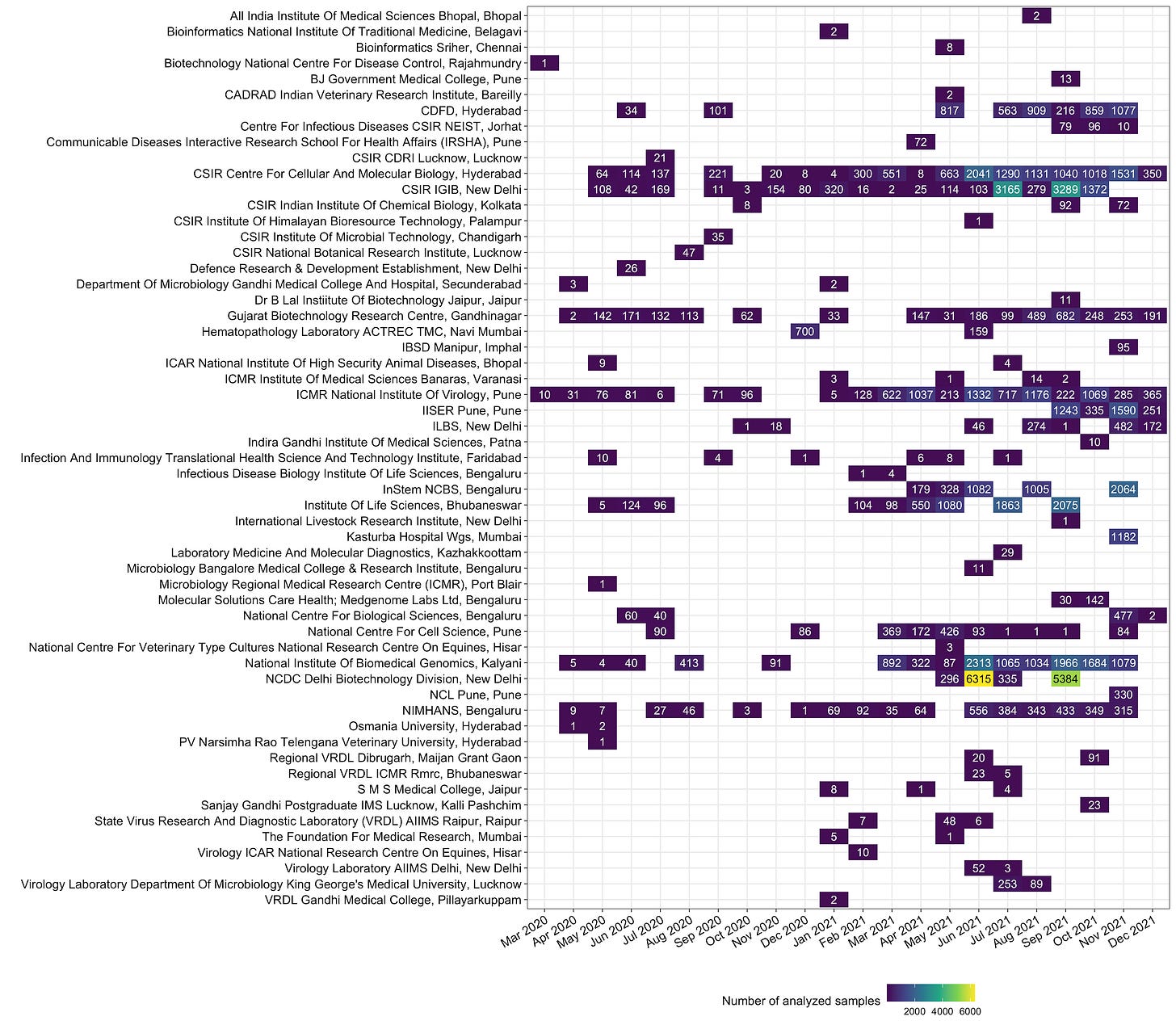
There is a huge disparity in the loads handled by the analysis labs. NCDC Delhi, has sequenced 12,330 samples, the highest so far. Together, NIBMG (Kalyani, WB), CCMB (Hyderabad) and IGIB (New Delhi), and NCDC have sequenced and deposited >50% samples (Figure 11).

INSACOG labs are not the only ones that have deposited sequences on GISAID. India commissioned a number of labs in 2020 that are currently not part of INSACOG, who also had sequences deposited on GISAID. This includes labs like CSIR National Botanical Research Institute (Lucknow) which sequenced 47 samples in August 2020, CSIR Institue of Microbial Technology Chandigarh which sequenced 35 samples in September 2020 or ACTREC, Tata Memorial Center, Navi Mumbai that sequenced 700 samples in December 2020, and 159 samples in June 2021. Given we have capacity beyond the core INSACOG labs, it would be worthwhile to consider all these other laboratories that have contributed in the sequencing efforts in the past to be a regular contributor.
Heavy lifting != efficiency: Long delays in uploading sequences to GISAID
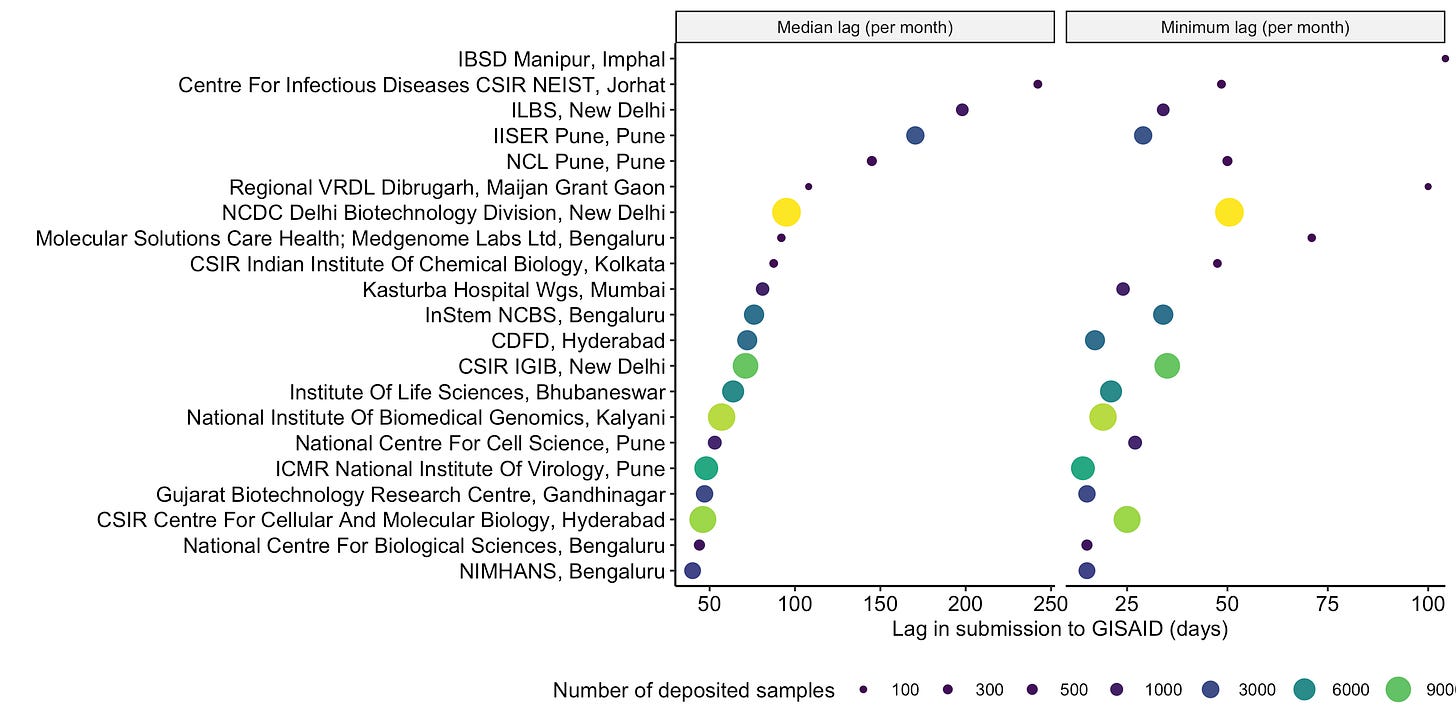
The first genomic sequences of Omicron (B.1.1.529) from Africa was uploaded on GISAID on 23nd November, a mere 7 days after it was collected from a sample in Gauteng, South Africa (the first sequence as of 3rd December 2021 on GISAID was from Hongkong which was uploaded on 22nd November and was originally collected on 13th November). Our labs have a median delay of ~ 70 days between when the sample is collected and before it shows up on GISAID. The median delay is strikingly different for each lab. For example. CSIR-CCMB deposits samples within ~45 days (median) of collecting them, while NCDC which has processed the highest number of samples so far takes ~120 days!
It is not clear what is the bottleneck in the entire workflow, but if we look at how quickly the samples have been deposited to GISAID after collection, over the entire timespan spanning the pandemic, even the samples with minimum turnaround time take ~30 days if they are analyzed at NCDC before it makes it to GISAID. When analyzed by looking at the median lag on all samples deposited in a month, this number remains largely unaffected (Figure 14), implying that the agency doing the heavy lifting is probably not as efficient.

The choice of sequencing technology
The labs in India mostly rely on Illumina or Oxford NanoPore technology for sequencing. While Illumina-based technologies require a proper sequencing facility set up (and hence more capital), the NanoPore technology is very “decentralized” - you can carry it like a USB stick anywhere, plug it into a laptop, and sequence the samples on the go. This can be a speedy and cost-effective alternate for academic labs that are interested in contributing (and are given access to necessary resources). For example, NCL Pune, BJ Government College Pune, NIHANS Bengaluru are a few labs that have made use of NanoPore technology in the recent past.
Unclear what our sequencing capacity is, but our public labs might be near saturation point
If we use the lag between samples collection and when it is submitted to GISAID as a proxy to define the sequencing capacity of each lab, around 172 samples (median) are analyzed every week. For example, NCDC is capable of sequencing >1500 samples per week based on its per-week submissions to GISAID so far. Past claims by the government have estimated the total sequencing capacity of the country to be 50, 00 samples per month.
The current capacity of the country is to sequence over 50,000 samples per month; earlier it was approximately 30,000 samples.
With 28 labs, this boils down to approximately 450 samples every week per lab. Based on the sequences deposited to GISAID, only NCDC Delhi, Kasturba Hospital Mumbai (which deposited 1000+ samples in a single month), and NIBMG Kalyani (WB), NCBS Bengaluru seem to have the capacity to process >450 samples per week. While not all the labs need to have a high capacity, adding more labs with even a bit smaller capacity would increase the overall capacity.
Leveraging our Private sector for ramping up surveillance
An earlier order from ICMR had banned private labs from performing any genomic sequencing. A lot of private labs not only have the infrastructure to do the genomic sequencing but can also sequence close to 200 samples every week. From a report in the Scroll:
Many of the bigger privately-owned networks of diagnostic laboratories in the country such as Strand Life Sciences and Metropolis have sequencing capabilities. So do some smaller stand-alone labs such as the Bengaluru-located Hybrinomics Life Science and Diagnostics. Prabhu Meganathan, who heads the lab, said they could “sequence “30-40 [sequences] per day” but “unless and until the government allows us to, we can’t”.
In September 2021, the Government decided to ease some of the restrictions and invited Private laboratories to be a part of INSACOG. Quoting a report from ET:
The biotechnology department has held discussions with private laboratories including New Delhi-based Mahajan Imaging and Premas Life Sciences, Bengaluru-based Strands Life Sciences and Genotypic Technology, Gurgaon-based NMC Genetics India, and Hyderabad-based Mapmygenome India.
This is a welcome decision. The private sector was instrumental in ramping up the RT-PCR-based testing in India and has untapped potential that we can leverage for ramping up genomic sequencing. In The US, a number of private corporations work closely with CDC and is partly responsible for the improved surveillance in the past few months.
A short report from the NITI Ayog on engaging private players to improve disease surveillance in India starts with a fantastic statement, that summarises my point:
It may appear quite bewildering to an outside observer as to why our National Health Programmes remain steadfastly shy of engaging with the private health care providers even though they havean overwhelming share in the provision of health care services. With an80% share in outpatient and 60% in inpatient care, 1 it is one of the highest proportions in the world, including developing economies. The default mode is to place reliance upon public sector health facilities- perceived as being exclusively in the Government domain, and therefore inherently more trustworthy – and, to some extent on not for profit organizations….
Wherever an attempt has been made to involve the entire health delivery mechanism public and private- the public health goal achievements have been far superior as in the case of Polio Campaign (eradication achieved) or in the Public-Private Mix (PPM) approach for Tuberculosis control (significantly improved case detection)

Interestingly, at least one private lab in India already seems to be contributing. I was not able to locate the lab of 28 lists under INSACOG, but based on the latest data uploaded to GISAID, Molecular Solutions Health Bengaluru / MedGenome have uploaded a total of 172 sequences in September and October 2021. Hopefully, we can involve more players in the coming weeks and expand our surveillance to capture Omicron prevalence. It probably would not end with Omicron, the virus will keep mutating by its virtue - we really need to catch up before it does (again!).

From the Twitterverse









