Genome #1: The state of Covid-19 genomic surveillance in India - Part 1
India needs a robust genomic surveillance plan. In Part 1 of a three parts series, we discuss the across-state disparities in genomic surveillance.
This post is Part 1 of a three-part series. In Part 2, we explored the variation in deposited samples across states, and in Part 3, we look at how involving private players can enhance our sequencing capability with minimal investment.
Covid-19 is now a preventable disease - vaccines work! Yet, the SARS-CoV2 virus continues to mutate and poses a continuous threat to public health. The delta variant, which is now known to be highly transmissible, had its role in the massive surge of cases that India witnessed during the second wave collapsing the healthcare system. In the US, where the mask restrictions were lifted for the vaccinated, the highly transmissible variant has forced CDC to reconsider its decision.
On 25th December 2020, the Government of India decided to set up the Indian SARS-CoV-2 Consortium on Genomics (INSACOG) to start genomic surveillance for Covid-19 in India with the following motivation:
In order to fully understand the spread and evolution of the SARS CoV-2 virus, and to tackle its future spread sequencing and analyzing the genomic data of this novel corona virus would be required. The study of accumulated of mutations in the viral genomes will enable us to compare virus samples and viral lineages in order to understand if local outbreaks are caused by transmission of single or multiple viral lineages. Analysis of SARS-CoV-2 genome sequences would also allow us to study the evolution of the virus and assess whether these mutations influence transmission, clinical outcomes, severity, or if they may impact interventions such as public health intervention measures and vaccines. Against this background, the sudden outbreak of a new SARS-CoV-2 variant in the UK requires India to increase viral Genomic surveillance in order to understand the spread of the virus in a rapid and robust manner.
What is Genomic surveillance?
Genomic surveillance involves sequencing a certain proportion of samples (swabs) that test positive for Covid-19 on an RT-PCR test. But, sequencing ≠ RT-PCR. Genome sequencing involves deciphering the order of nucleotides that make up the RNA of the virus. In contrast, the RT-PCR test can only detect the presence of a virus but does not provide enough information about its sequence (variants).

What would one learn from such a permutation of just four alphabets (bases)? The RNA of SARS-CoV2 is made up of ~30,000 bases. Once it infects human cells, the coronavirus tries to make copies of itself by replicating its RNA. During this replication, the machinery responsible for copying which is inherently error-prone can introduce mutations or changes. Most of these mutations are of low significance for the virus, allowing it to replicate as it would otherwise. But some mutations can have stronger biological effects, both for the virus and humans. Certain mutations in the virus can alter its infectivity, disease severity, and how it interacts with our immune system or the vaccines. Genomic surveillance allows us to examine and compare the genome sequence of the viral strains infecting the population. By characterizing the mutations in the sequence, it is possible to understand how quickly a strain is spreading. its association with (increased) transmission and its likely impact on the existing vaccines. Hypothetically, if the original SARS-CoV2 virus had its genomic sequence as ATTAA, a variant sequence might be ATTAC, probably making this variant more severe in its attack on the respiratory system. For example, characterization of Delta variant and follow-up studies have characterized that people infected with Delta variant carried 1000x more viral load, thereby leading to its increased infection rate. Luckily, vaccines remain effective, though the mutations are potentially less susceptible to some of the treatment strategies currently in use. To be better prepared, we need to stay one step ahead of the virus and genomic surveillance at scale enables exactly that.
The role of INSACOG in Genomic surveillance
India took a whole 11 months since the first case was detected in the country to formally set up a dedicated consortium to perform surveillance. Though, national laboratories had started sequencing samples as early as April 2020, when around 4,000 samples were sequenced and deposited to GISAID, an open repository to submit influenza and SARS-CoV2 genomic sequences. On the other hand, United Kingdom (UK) had a consortium established with ample funding as early as April 2020. No doubt, UK has also emerged as a leader with around half a million sequences deposited on GISAID. In contrast, India has had only ~36,000 sequences deposited as of July 20th, 2021, just 0.116% of the total infections detected in the same time frame.
The government did have a specific goal in mind for INSACOG - sequence 5% of the samples testing positive for SARS-CoV2 as the B 1.1.7 became a variant of concern:
Selection of samples for genome sequencing:
Representative positive specimens ( randomly sampling 5% of the positive specimens) – approximately 5% of the specimens detected positive since 23rd November, 2020 will be sent to the designated Regional Genome Sequencing Laboratory (RGSL) (to be provided by CSU in discussion with the respective sate SSOs) – details of the cases reported from various States since 23 November, 2020 is in the annexure 4. It is pertinent to mention that there have been variations in the States/districts with regard to prevalence and incidence. Prioritization can be done for COVID labs/hospitals in urban regions and tertiary care health facilities at district level. Hence, CSU will ensure a proper representation particularly from the metro cities where there is high probability of importation of new SARS-CoV-2 variant.
It is also worth highlighting one of the challenges enlisted in the guidance document:
The genome sequencing for assessing the current status from the samples tested positive during past months (September 2020 onwards) will depend on the availability of aliquots of positive samples in Government/private labs as, it has come to the notice that many of the labs are not storing the positive samples. While specimens collected in the past will have such limitations, prospectively collected specimens can be used on a weekly basis for 5% random draw based representative sampling.
The choice of 5% though may appear random, has been shown to be optimal using a mathematical in a recent study uploaded on medRxiv preprint server without peer review and funded by Illumina, a biotechnology company. It recommends at least 5% of samples testing positive should be sequenced if the prevalence rate is between 0.1-10% (WHO also recommends around 10% of samples be sequenced):
By modeling common performance characteristics of available diagnostic and sequencing tests, we developed a model that assesses the sampling required to detect emerging strains when they are less than 1% of all strains in a population. This model demonstrates that 5% sampling of all positive tests allows the detection of emerging strains when they are a prevalence of 0.1% to 1.0%. While each country will determine their risk tolerance for the emergence of novel strains, as vaccines are distributed and we work to end the pandemic and prevent future SARS-CoV-2 outbreaks, genomic surveillance will be an integral part of success.
India’s sequencing efforts rank lower than other nations with similar economic status
Countries like New Zealand, Australia, and Iceland have been able to have >50% (as high as 75% in the case of Iceland) of their Covid19- cases sequenced and shared while maintaining the total deaths to under 10 per 100,000 people (Fig. 1A). It can be argued that these countries could do large-scale surveillance since they are richer, with a per capita GDP exceeding $25,000 (US Dollars) while compared to India’s $2,000. But even if we account for the difference in GDP, India performs poorly, both in terms of the total number of deaths per 100,000 and the percent of sequences shared and sequenced (Fig. 1B: compare with Bangladesh and Zimbabwe for example, with similar GDP yet higher percentage of sequencing with lower deaths overall).
In all three countries, genomic surveillance has been used to drive the policies. For example, Australia and New Zealand by sequencing most of its Covid-19 cases keep track of variants of concern, pinpoint the origin of the outbreak, and uses it to impose lockdowns - Science-backed lockdowns. On the other hand, Iceland has been less stringent about imposing lockdowns given that its economy heavily relies on tourism, yet by keeping up with genomic surveillance has been largely successful in containing the spread of the virus.
India has sequenced only ~0.12% of all Covid-19 cases
India’s tally for sequences shared on GISAID stood at 36,450 as of July 20th, 2021. The elusive denominator, in this case, is a massive 31.5 million cases (at least) that India has detected so far (Fig. 2). This brings down the national percentage to a mere ~0.12%. To reach the suggested 5%, we will need to ramp up our sequencing capacity by at least 450 times! With 28 labs (only 10 are currently listed, the identity of the remaining 18 remains unknown) currently part of INSACOG, this roughly boils down to each lab sequencing at least 15 times more samples than their current status.

Variation across states
In its guidance document, INSACOG points out
It is pertinent to mention that there have been variations in the States/districts with regard to prevalence and incidence.
There is no doubt, that this is indeed the case. However, there is huge variation in the representation of sequenced samples across states (Fig. 3). For example, two states, Maharashtra and Kerala, that have always been a matter of discussion (and often concern) for their cases have disproportionately different percentages of samples sequenced and shared on GISAID. Maharashtra which has been seeing a median of 2,03,518 cases per month (non-cumulative) has deposited 0.114% of its cases overall. Kerala, on the other hand, with a median of 1,30,225 cases per month only has deposited 0.0023% (median) of its cases every month which is next to Bihar’s tally of 0.0004% for its median 15,153 cases. Kerala’s low sequencing rate might be partially explained by the fact that the sequences are being separately tracked through another consortium, though the raw data and metadata remain inaccessible on the portal.
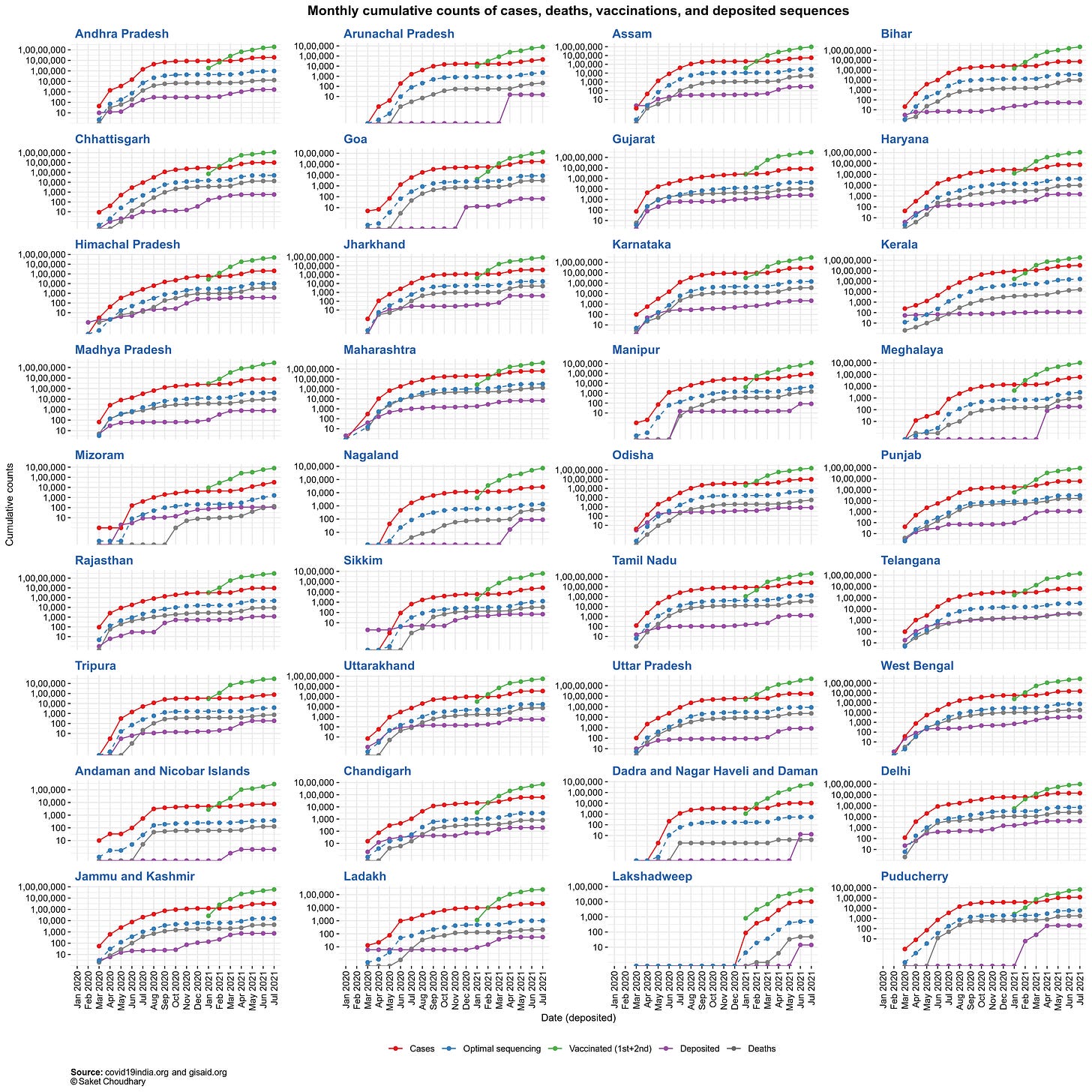
Surprisingly, India was able to sequence an optimal number of samples across states at the beginning of the pandemic, when INSACOG did not exist. Barring the March-April 2020 and Feb-April 2021 periods, the nationwide and statewide sequencing proportions remain low (Fig. 4) while only a handful of states contribute significantly towards driving the national average higher, of particular note being Telangana with a median sequencing of 0.83% samples (Fig. 5), which is 0.71% additional over the national average of 0.12%.


“Could we have prevented this?”
The initial set of 10 labs that constitute INSACOG have the capacity to process 30,000 samples a month. Hypothetically, if India registers 50,000 Covid-19 cases every day for a month (or 15,00,000 cases per month), we will need to sequence 75,000 cases every month - stretching the current capacity by an extra 45,000. But even with its current capacity, the current count of deposited sequences on GISAID (~36,000) clearly indicates that the labs are not operating at their capacity (they do not need to necessarily). So the fact that we are still getting a lower percentage of sequences deposited could not be because of capacity saturation. If the labs have the capacity, and we are still not processing enough samples what is the point of failure in the system? One possibility is the scarcity of funding. If the Government cannot make funds available to ensure good public health, especially in the time of a pandemic, there is very little we could hope to change. A second possibility is the lack of good samples arising from the unavailability of proper reagents (resources) and trained staff to prepare and handle the samples. While the only solution to the first problem is ensuring enough funding, the second problem warrants alternate solutions that we will discuss in Parts 2 and 3 of this series. We will also highlight how not only we are not sequencing enough, there is also a significant delay in when the samples get collected (and likely sequenced) and when they get uploaded to GISAID - for example, the delta variant (B 1.617.2) could have been characterized as early as September 2020 (instead of December 2020) had the sequences been shared publicly in time.
The virus will not stop mutating, to make sure its spread does not spiral out again, we need a robust genomic surveillance program. The focus of the program should be on ensuring at least 5% of Covid-19 cases are sequenced and samples from across states have the required representation - something the INSACOG’s entire foundation is based upon.
First posted: July 27, 2021 - 10:00 AM EST.
Last updated: July 27, 2021 - 2:30 PM EST.







