Genome #3: The state of Covid-19 genomic surveillance in India - Part 2
Significant delays in genomic surveillance data analysis and submission
This post is Part 2 of a three-part series. In Part 1 of this series, we looked at how India’s genomic surveillance has been abysmally suboptimal, both at the national and state level, with a huge state-to-state variation. In this post, I analyzed the variants data deposited by India to GISAID and looked at some state-level metrics. In Part 3, we look at how involving private players can enhance our sequencing capability with minimal investment.
It is a capital mistake to theorize before one has data.
—Sherlock Holmes.
Till August, India has detected ~ 33 crores Covid-19 cases. In Genome # 1, we explained the importance of sequencing a proportion of these cases can help us understand which variants are prevalent and if arising new variants are growing at a rate that is concerning.
INSACOG, India’s SARS-COV-2 Genomic Sequencing consortium, is responsible for monitoring the genomic variations in SARS-Cov-2. INSACOG’s website has a section where updates are posted weekly, which look like this:

The idea behind summarizing this information is to highlight if any of the known variants are trending towards becoming a variant of interest or concern. Center for Disease Control and Prevention, describes a variant of interest as:
A variant with specific genetic markers that have been associated with changes to receptor binding, reduced neutralization by antibodies generated against previous infection or vaccination, reduced efficacy of treatments, potential diagnostic impact, or predicted increase in transmissibility or disease severity.
Possible attributes of a variant of interest:
Specific genetic markers that are predicted to affect transmission, diagnostics, therapeutics, or immune escape.
Evidence that it is the cause of an increased proportion of cases or unique outbreak clusters.
Limited prevalence or expansion in the US or in other countries.
As of September 5th, 2021, the following Pangolin lineages are of interest:
B.1.525 (Eta),
B.1.526 (Iota)
B.1.617.1 (Kappa)
B.1.617.3
On the other hand, variants of concern are defined as:
A variant for which there is evidence of an increase in transmissibility, more severe disease (e.g., increased hospitalizations or deaths), significant reduction in neutralization by antibodies generated during previous infection or vaccination, reduced effectiveness of treatments or vaccines, or diagnostic detection failures.
Possible attributes of a variant of concern:
In addition to the possible attributes of a variant of interest
Evidence of impact on diagnostics, treatments, or vaccines
Widespread interference with diagnostic test targets
Evidence of substantially decreased susceptibility to one or more class of therapies
Evidence of significant decreased neutralization by antibodies generated during previous infection or vaccination
Evidence of reduced vaccine-induced protection from severe disease
Evidence of increased transmissibility
Evidence of increased disease severity
Current variants of concern are:
B.1.1.7 (Alpha)
B.1.351, B.1.351.2, B.1.351.3 (Beta)
B.1.617.2, AY.1, AY.2, AY.3, AY.4, AY.5, AY.6, AY.7, AY.8, AY.9, AY.10, AY.11, AY.12 (Delta)
P.1, P.1.1, P.1.2 (Gamma)
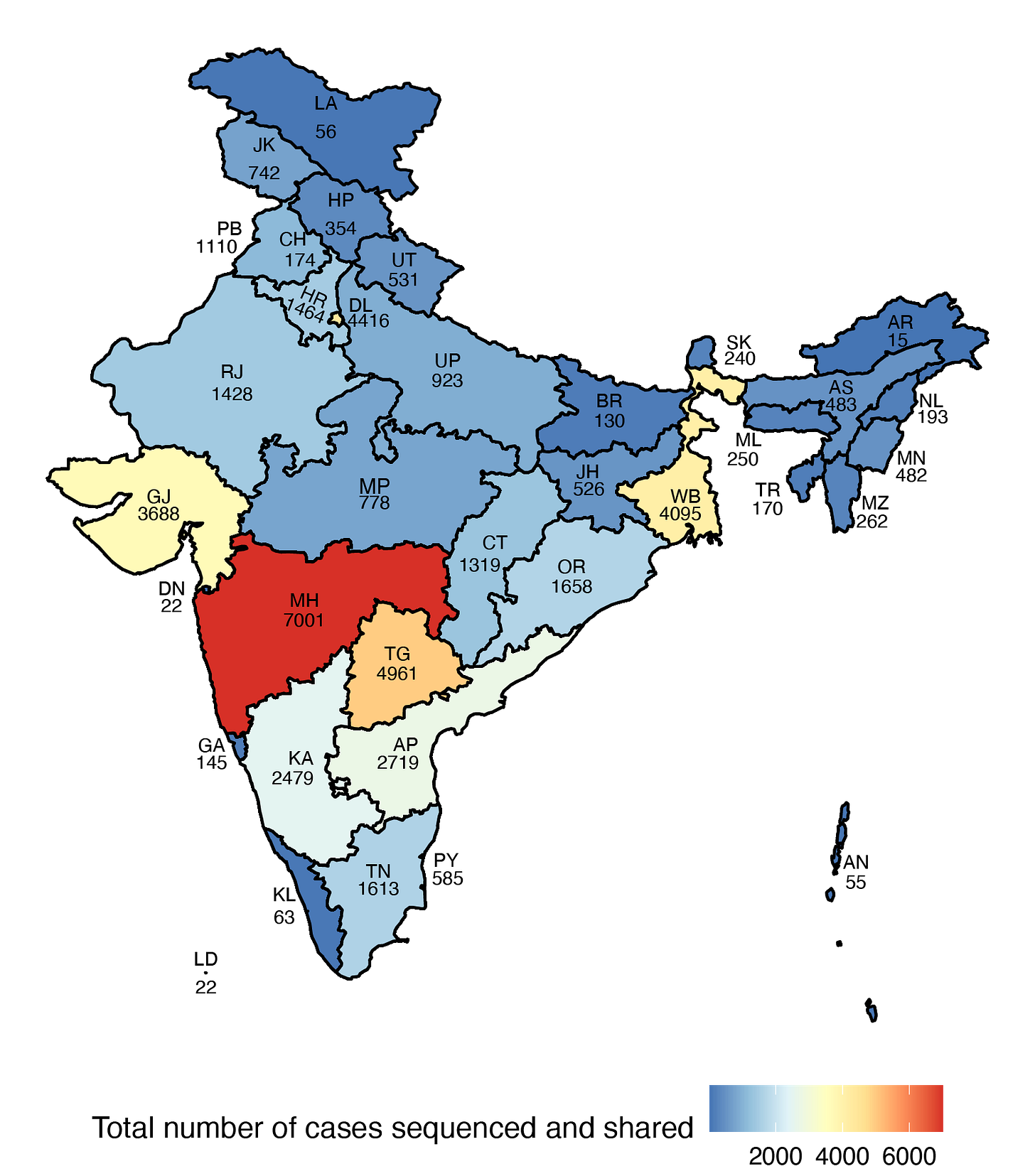
While INSACOG’s bulletin summarizes the total number of sequences, it is currently not possible to access the state-specific data. Its data portal has an API that has limited functionality and makes obtaining data harder than it should be. Part of this data also gets deposited to GISAID, an open repository where multiple countries upload SARS-CoV-2 genomic sequences. This data has state-specific granularity, which when visualized presents the following picture of the distribution of genomic sequences across states:
The state-specific data highlights the disparity in sequencing, but it is also expected given the case-load in each state was different to start off with. If we adjust for the total number of Covid-19 cases in each state, the percent of cases that are sequenced and shared is highly variable (Figure 3). While cases like Sikkim and Telangana have around 0.8% of cases that are sequenced and shared, Kerala has so far deposited only 0.001% of the 40+ lakhs cases it has witnessed so far.
One would imagine that sequencing a case, analyzing the data, and sharing them on GISAID should be a matter of a couple of weeks or a month at maximum (sequencing and data analysis is not the bottleneck here and in principle can be finished in a few days to a week). But data from GISAID deposited from India shows a huge disparity in the delay involved between sample collection and its deposition on GISAID. India has a delay of ~ 62 days (median) - it takes at least two months before a case detected and sequenced in India makes it to GISAID for the world to be informed! On a state level, these numbers have a varied range from as small as 28 days for samples collected from Dadra and Nagar Haveli to 120+ days in the case of Himachal Pradesh and Andaman & Nicobar Islands.

Why is the delay in submission to GISAID a problem?
In Part 1, we looked at how India (and almost every state) is way behind the sequencing target of 5% (for tracking the variants in a robust fashion, it is recommended to sequence 5% of Covid-19 cases). However, not sequencing enough is not the only problem. Assuming the delay in submitting to GISAID involves a delay in analyzing these sequences even at the INSACOG end, this might give enough time for a new variant to spread before any sort of preventive (policy) action can be taken. A study in the New England Journal of Medicine highlights that SARS-CoV-2 with multiple mutations can arise in people with immunocompromised (those with HIV/AIDS, cancer; on immune system suppressant drugs; or inherited diseases that affect the immune system).
Patients with immunosuppression are at risk for prolonged infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In several case reports, investigators have indicated that multimutational SARS-CoV-2 variants can arise during the course of such persistent cases of coronavirus disease 2019 (Covid-19).1-4 These highly mutated variants are indicative of a form of rapid, multistage evolutionary jumps (saltational evolution; see Glossary), which could preferentially occur in the milieu of partial immune control
This is a key challenge for India given the huge burden of the immuno-compromised population (for example, India sees 13.5 lakh cancer cases every year).
How can early submission of genomic surveillance data help us?
WHO declared Delta (B 1.1617.2) as a variant of concern on May 11th 2021:
Assuming India deposits data on GISAID in real time or within a week, it would enable flagging variants of interest well ahead of time. For example, I looked at only samples collected between January and March 2021 and compared the growth rate of all variants to B 1.1.7 (alpha) which was in abundance in the preceding months. It is hard to predict how much would have changed with this information, but significant delays in data submission do open holes for capital mistakes, as Sherlock would say.

An effective genomic surveillance system should target not only optimal sequencing (5%) but also that these are analyzed and submitted to public repositories like GISAID which can not only enable early characterization of its biological consequences but also pave the way for effective public policies well in time.